Technical support
What kind of technical support do we provide?
Euro-BioImaging offers technical assistance to users in the form of tool and workflow development which facilitate FAIR image data management and analysis. To this end, a particular focus is laid on developing tools that support :
- The preclinical imaging community and promote open science and FAIR practices by providing tools for annotation and upload of preclinical datasets:

XNAT-PIC: The XNAT for Preclinical Imaging Centers (XNAT-PIC) is a free and open-source Windows desktop application, which offers several tools to expand the XNAT core functionalities to. The app facilitates annotation and uploading of image datasets to the XNAT platform, focussing on the specific requirements and structure of preclinical imaging studies .
- The Next Generation File Format (NGFF), a community-supported file format intended to fulfil the FAIR standards for image data. Importantly, NGFF offers various advantages with respect to data access, management and analysis, which are missing in most traditional, TIFF-based formats. A specific implementation of NGFF is OME-Zarr, which is a layer on the chunked file format Zarr.
Why are we supporting the OME-Zarr format?
OME-Zarr permits multidimensional chunking of image data, a feature that enables efficient streaming of arbitrary subsets of data. Unlike most traditional file formats that are limited to plane-based access, the multidimensional chunking enables faster access to complex multidimensional data stored in the OME-Zarr format. OME-Zarr also offers native support for multi-scaled, pyramidal image datasets, a feature that dramatically improves the performance of image data visualisation, especially with datasets stored in object or cloud storage. A comparison of access performance between OME-Zarr and some other popular image file formats can be found here. In addition to these performance benefits, OME-Zarr also provides community-supported layouts and specifications for rich data representation, which inherently supports multiresolution images, their core metadata as well as segmentation masks and certain types of image analysis results in the same pyramidal hierarchy. To fully harness the advantages of the OME-Zarr format, the community is building a range of tools around it, leading to an OME-Zarr ecosystem. A non-exhaustive list of these tools can be found here.
What types of tools are we developing?
- Support for image data conversion to open and accessible file formats
Euro-BioImaging strives to equip users with tools that facilitate image data conversion to open and accessible file formats, and in particular, enable a swift initiation into working with the OME-Zarr format. BatchConvert is a command line interface capable of concurrent conversion of image data collections from various proprietary file formats to either of the standard formats OME-TIFF or OME-Zarr, utilising the workflow management tool Nextflow. BatchConvert offers the flexibility of coupling conversion with the concatenation of multiple images along specified dimensions. An important feature of BatchConvert is that it is capable of automatic job submission on Slurm-managed clusters. The same conversion command can be run on a local machine or on an HPC cluster depending on the specification of a single command line argument. This feature makes BatchConvert readily incorporable into multi-user HPC environments. BatchConvert is also designed to support remote input and output, meaning that it can optionally import data from s3 buckets for conversion and can export the output back to s3 buckets, hence coupling conversion to data transfer. The remote IO support extends to the capability to automatically submit the converted data to the BioImage Archive, which streamlines the process of making scientific image data open and accessible. The GitHub repository of BatchConvert holds the source code as well as some documentation and examples for its use. The recommended way to start using BatchConvert is to install the Anaconda package. To learn more about BatchConvert, please also have a look at our preprint.
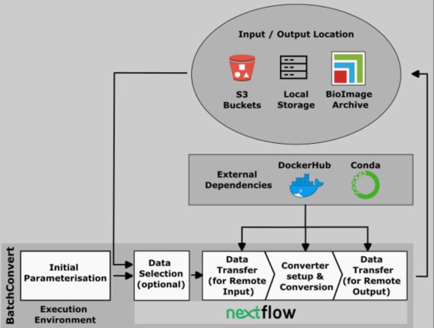
Outline of the BatchConvert workflow
- Library support for reading, writing and processing OME-Zarr datasets
To facilitate the processing of image data stored in the OME-Zarr format, we are developing a Python library, namely, ome_zarr_pyramid, which enables reading, writing as well as modifying OME-Zarr datasets from a Python environment. Ome_zarr_pyramid provides a Python object that leverages the OME-Zarr pyramidal layout to internally represent an image, and provides a wide range of methods and functions that can process this OME-Zarr object. Any modifications to the data, such as change of voxel type, splitting/subsetting of the image array, deletion/addition of resolution layers, etc., introduced by these methods and functions, are automatically handled by the library and incorporated into the output OME-Zarr’s layout. To extend ome_zarr_pyramid, two approaches are being used: i) by providing OME-Zarr support to existing image processing libraries which are currently limited to other file formats, ii) by providing custom functionalities.