Euro-BioImaging’s EMBL Node contributes to understanding SARS-CoV-2 replication cycle in human cells
Understanding virus replication is a key part of any therapeutic strategy to combat COVID-19. Without this crucial piece of the puzzle, it is hard to develop drugs to suppress viral replication and virus-induced cell death. EMBL’s Electron Microscopy Core Facility, part of Euro-BioImaging’s EMBL Node, participated in a timely study to reveal the biological mechanisms driving the SARS-CoV-2 replication cycle in human cells. This study, carried out in collaboration with the Ralf Bartenschlager Lab, of the Department of Molecular Virology at the University of Heidelberg, and the Schwab team, of the Cell Biology and Biophysics Unit at the EMBL Heidelberg, combines high-throughput imaging technologies, a massive volume of images, and extraordinary working conditions to create 3D reconstructions of whole cells that are infected by SARS-CoV-2 and their subcellular compartments. The impressive datasets produced in this study are publicly available via the EMPIAR database (ID 10490), giving scientists new insight into the way this novel virus behaves within human cells. Yannick Schwab, Head of EMBL’s Electron Microscopy Core Facility and team leader in CBB, explains the contribution of the EMBL Node to this important study, “Integrative imaging reveals SARS-CoV-2 induced reshaping of subcellular morphologies,” published in Cell Host & Microbe.
State-of-the-art imaging techniques
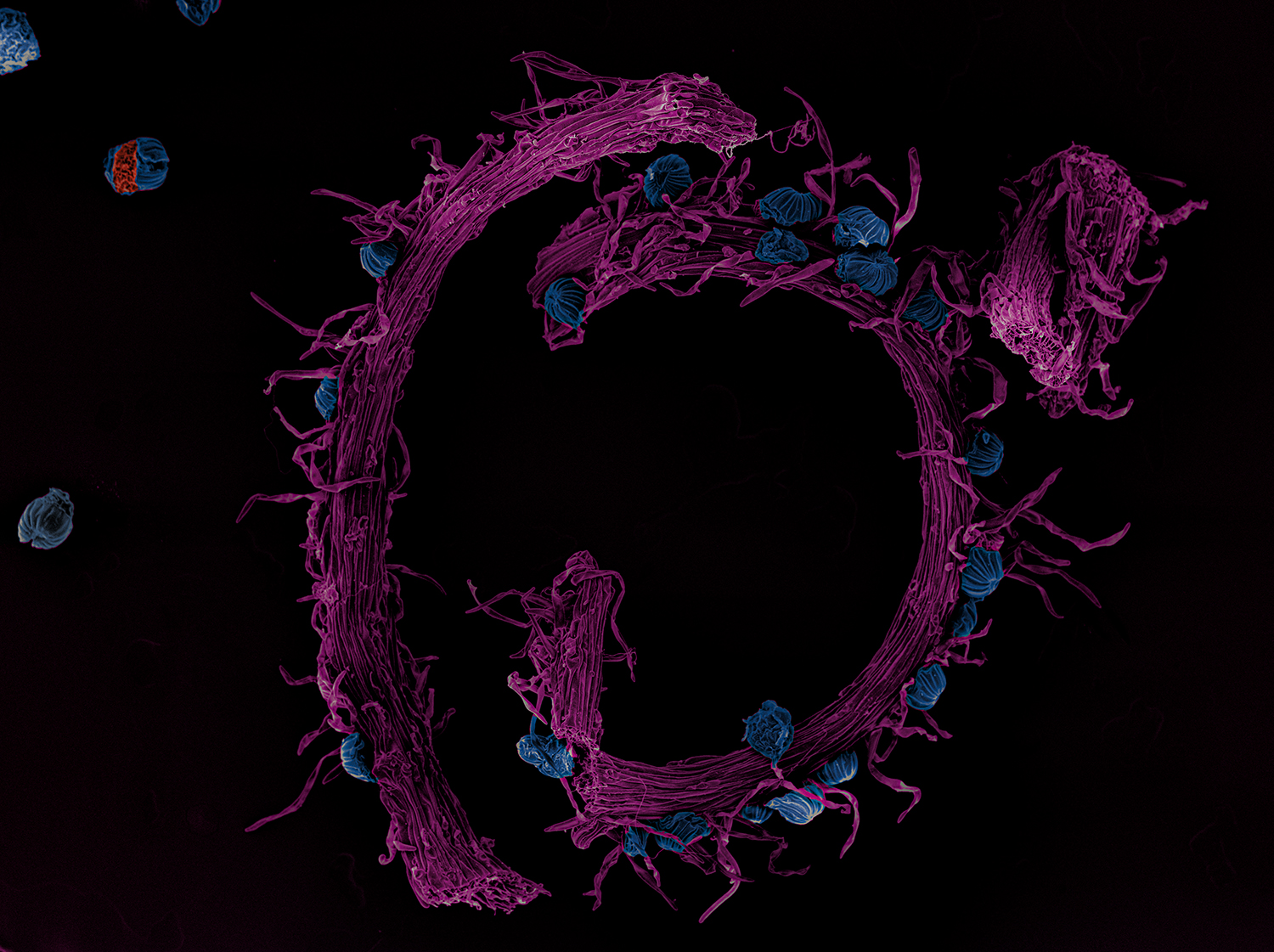
Yannick Schwab: This study combines light and electron microscopy and image analysis to provide a comprehensive overview the morphological organelle alteration induced in SARS-CoV-2 infected human lung epithelial cells. The expertise of the Bartenschlager lab for sample preparation in Biosafety level 3 conditions and light microscopy image acquisition and analysis, combined with our own expertise in Electron Microscopy (EM) image acquisition, analysis and data sharing, were central to this study. Our contribution was to use different EM techniques to visualize the steps in the virus replication process, including using FIB-SEM to study full cells in 3D at high resolution. We applied automated and manual techniques to segment infected cells into color-coded organelles, making it possible to visualize virus replication at 6h, 12h and 24h post-infection. We could see the virus using the Endoplasmic Reticulum (ER) to create replication factories in double membrane vesicles (DMV) and the new virions forming at the ERGIC/Golgi interface. To gain insight into the complex subcellular events induced by the infection, we reconstructed their 3D architecture by high resolution electron tomography.

Right: FIB-SEM imaging reveals in 3D the ultrastructure of an entire infected cell. Credit: Julian Hennies, Schwab team, EMBL.
For each tomogram, we identified and classified the cellular and viral structures present. This large dataset allowed us to identify different topological compositions of the SARS-CoV-2 induced organelle perturbations.
Our observations were consistent with other recent reports, confirming that SARS-CoV-2 alters the ER network to generate DMVs similar to other betacoronaviruses such as SARS-CoV-1, MERS-CoV and MHV. But beyond the similarities, we also observed some interesting alterations specific to SARS-CoV-2 that need to be studied further. Amongst the most striking are the formation of ER connectors that link the DMVs to the endoplasmic reticulum.
A full description of the techniques we used and what we observed are published in Cell Host & Microbe, “Integrative imaging reveals SARS-CoV-2 induced reshaping of subcellular morphologies”.
Extraordinary imaging analysis capacity
This study required an enormous amount of resources for image acquisition and analysis, which would not have been possible without the support of EMBL’s IT services and the extreme motivation of the EMCF and Schwab team. From April 29th- May 25th, we acquired nearly 10,000 images from our JEOL 2100 Plus electron microscope, adding up to close to 95 hours at the bench. These images were used to make 3D tomograms. In total, we reconstructed 243 dual-axis tomograms, manually segmenting high resolution images, representing an additional 140 hours transferring and registering grids to our ThermoFischer TF30 microscope.
Sharing one of the largest biological datasets ever created
With 5 FIB-SEM volumes and 244 high-resolution TEM images available to the scientific community for download through the EMPIAR platform, this study possibly provides one of the largest shared biological datasets ever. So other scientists can visualize the datasets smoothly, we are using MoBIE, an open access visualization tool, developed by Christian Tischer of the Center for Bioimage analysis at EMBL.
An extremely dedicated team
When we were first contacted by Ralf Bartenschlager in March to collaborate in this study, our lab was closed due to the lockdown. We had to request special permission to open the lab and all work on-site had to be done on a voluntary basis. Despite this context, everyone in my team and facility wanted to participate in this project. EMBL’s IT services showed great dedication in giving us external access to microscopes and providing computing services in the cloud. And Zeiss supported our study by sending an engineer to our facility to service our FIB-SEMs, despite the lockdown. Thanks to this overwhelming enthusiasm, we were able to do a year’s work in just a few weeks' time.
About Euro-BioImaging’s EMBL Node:
The Euro-BioImaging EMBL-Node offers a collection of state-of-the-art microscopy equipment and image processing tools. This Node supports in-house scientists and visitors in using microscopy methods for their research and regularly organizes international training courses to teach basic and advanced microscopy methods. The services provided include project planning, sample preparation, microscope selection and use, image processing and visualization. Through Euro-BioImaging, life scientists, regardless of their affiliation or area of expertise, can apply to access services from 21 internationally-renowned imaging facilities called Nodes.
More news from Euro-BioImaging